Understanding the Topology of Corona Literature
This analysis seeks to understand how the COVID-19 academic literature is organized. Do certain articles come together, and can these groups help identify key literature for the Covid-19 epidemic? To answer this question, this analysis will utilize self-organizing maps (SOM) to create clusters of academic articles. This analysis will be broken down into three sections: section one will discuss the methodology of creating COVID clusters; section two will show the results from training; and section three will give closing remarks.
Important links for this analysis:Methodology:

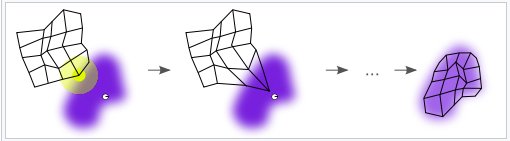
The data for this analysis was collected from a Kaggle competition, and it consists of over 29,000 academic articles and abstracts which relate to COVID -19. Due to limits in processing power, only the abstracts from each paper was analyzed. The processes for analyzing the COVID clusters can be found in the flow diagram above.
The first step to analyzing the COVID papers was acquiring the data from Kaggle. After acquiring the data from Kaggle, a CSV file was created which had each observation as a COVID article with main text and abstract as features. The proceeding steps in the process map were cyclic and were repeated a total of five times.
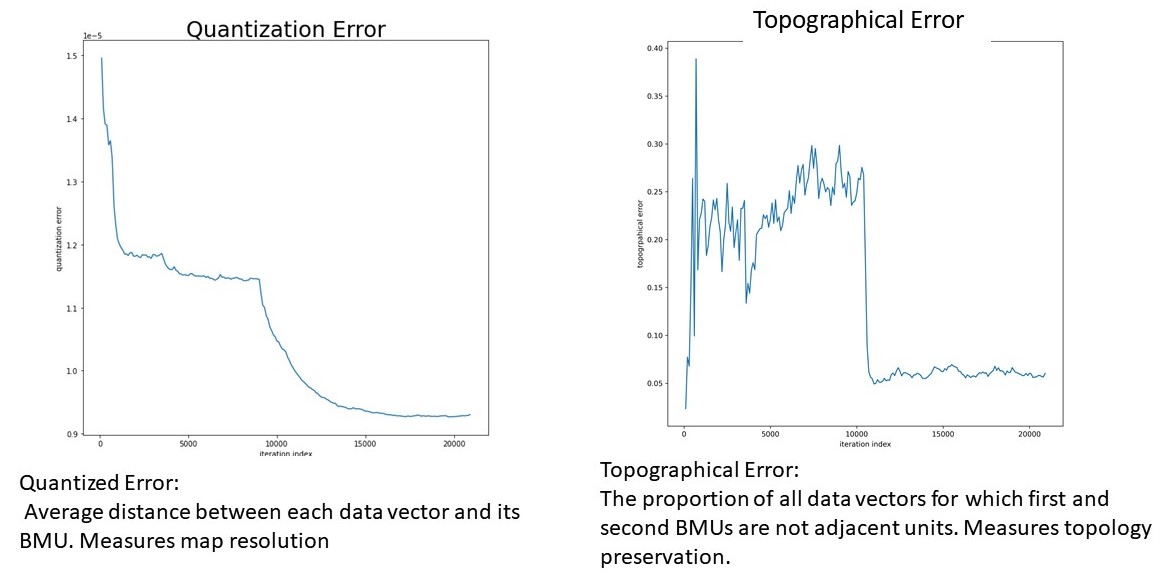 The next step took each article and created a term frequency-inversed document frequency L1 normalized matrix (TFiDF). The parameters for making the TFiDf consisted of keeping one and two word ngrams, stemming, and removing English stop words. After creating a TFiDf, the training process for the self-organizing map was started. At every run, the SOM was initialized with PCA weights, and every run ran through each observation once. As demonstrated in the figure below, at every training phase topographical and quantized error was checked in order the ensure that the SOM converged to a steady-state.
The next step took each article and created a term frequency-inversed document frequency L1 normalized matrix (TFiDF). The parameters for making the TFiDf consisted of keeping one and two word ngrams, stemming, and removing English stop words. After creating a TFiDf, the training process for the self-organizing map was started. At every run, the SOM was initialized with PCA weights, and every run ran through each observation once. As demonstrated in the figure below, at every training phase topographical and quantized error was checked in order the ensure that the SOM converged to a steady-state.

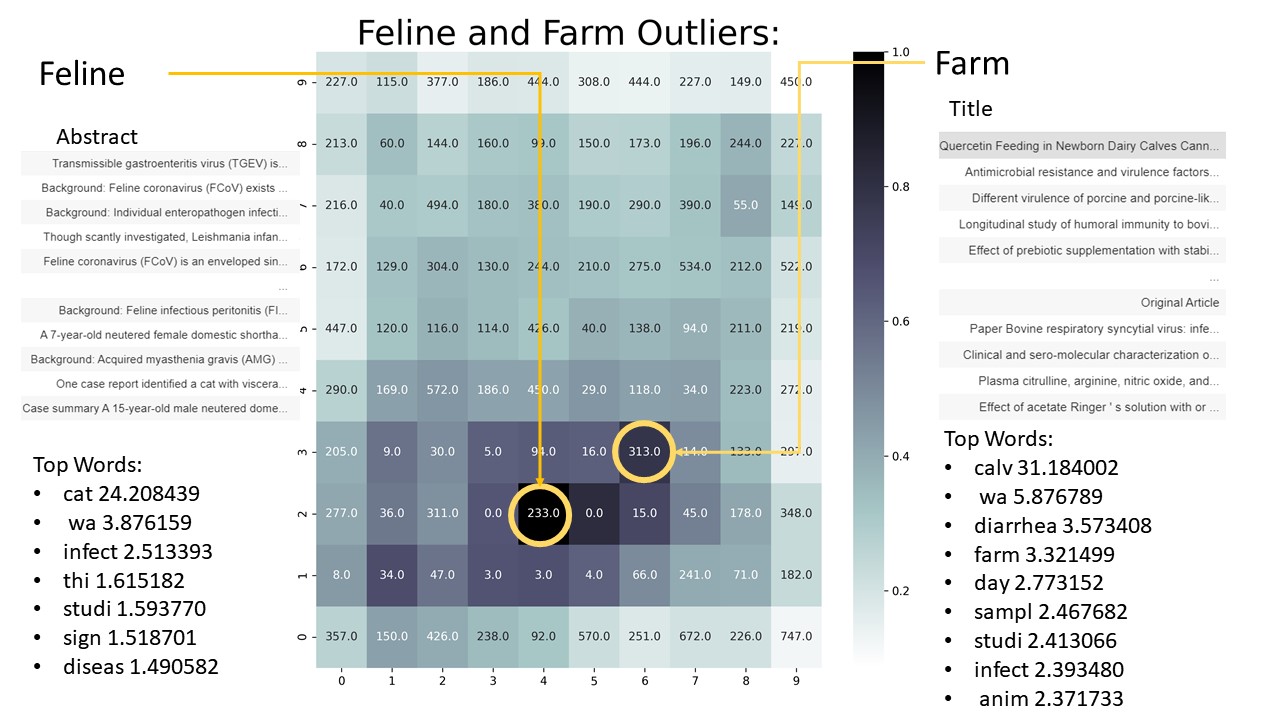
After each training phase, the results from the SOM were analyzed. The SOM was analyzed by looking at the distances between neurons, and by the number of observations each neuron had. An example of the graph used for this analysis can be found to the right. Here, each neuron is represented by a square, and the color of the square represents the average distance between neurons. The count of each neuron can be found in the center of each square. For the most part, no neurons that had a distance of less than .7 or had less than 100 observations were considered for further analysis. The neurons which were chosen for further analysis were analyzed through word frequencies and title context. After the analysis, these neurons were dropped, and the process was started again. This process eventually ended due to a lack of discernable differences between neurons.
Results:
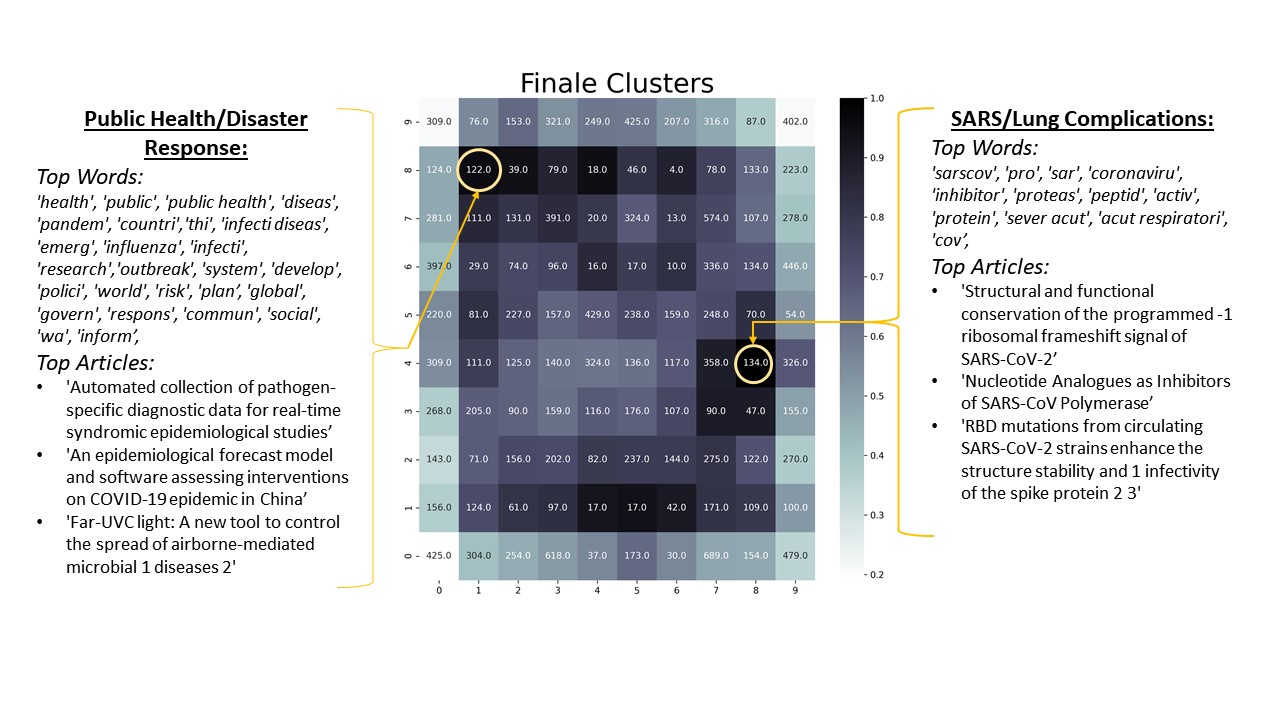
The next five figures will look at the graphs for each SOM, and to the left and right of each graph word frequencies and top titles can also be found. Word of caution, I do not have any type of medical or biological background, and therefore I lack the domain knowledge to draw any meaningful conclusions. the results from this analysis are merely knowledge for its own sake.



Closing Remarks:
As shown in the results above, noticeable clusters formed out of the COVID literature. Not only did meaningful clusters form, but local distances also seemed to be preserved. Related neurons seemed to be found near one another. Some neurons that are related and close to one another include: the Cell and Protein cluster is near the DNA and Genome cluster; the MERS cluster is near the Bats cluster; and the Feline cluster is near the Farm cluster. In conclusion, it can be said the there is an underlying structure and organization to the COVID literature. Understanding this structure could help parse out meaningful articles. For example, knowing which articles deal with public health could help identify which articles researchers need to read. Since local distances were preserved, neurons in-between the different clusters could show a mix and match of previously defined clusters. For example, in between the MERS and Bats cluster, I would expect to find articles that deal with the transmission of MERS.